10. Microbiome
Find a research or journal article where researchers cultivate 2 or more microbial strains. What technology are they utilizing? How scalable is this approach to more than 2 strains? How do they address issues related to requiring multiple media?
Microbial biophotovoltaics (BPV) are an exciting solution for renewable clean energy production. A biological alternative to solar power generation - the idea is to use biological photosynthetic materials and/or organisms to convert solar energy to electricity. Previous research has been done on using a mono-culture of photosynthetic microorganims, such as cyanobacteria or algea, to act as a BPV. Conventional BPV systems fix the organism cells to an anode to mediate the electron transfer outside to the cell. This process is apparently inefficient, or at least not well understood.
The researchers in the above paper offered and implemented a unique approach. BPV system basically requires two steps - charging (photosynthesis) and discharging (extracellular electron transfer). Therefore, they created a microbial consoritum system, where one species is in charge of charging (pun intended) and a second species is discharging. They had to figure out which species to use, as well as how to transfer the energy between the species. They chose to use an engineered cyanobacterium (Syn2973-omcS-ldh) as the light harvester. This strain is equipped with both photosynthetic machinery and D-lactate synthesis pathway. This D-lactate is the electron carrier to the second species - an engineered Shewanella oneidensis MR-1 (S. oneidensis-ΔnapA). This strain is equipped with D-lactase oxidation pathway and the extracellular electron transfer machinery.
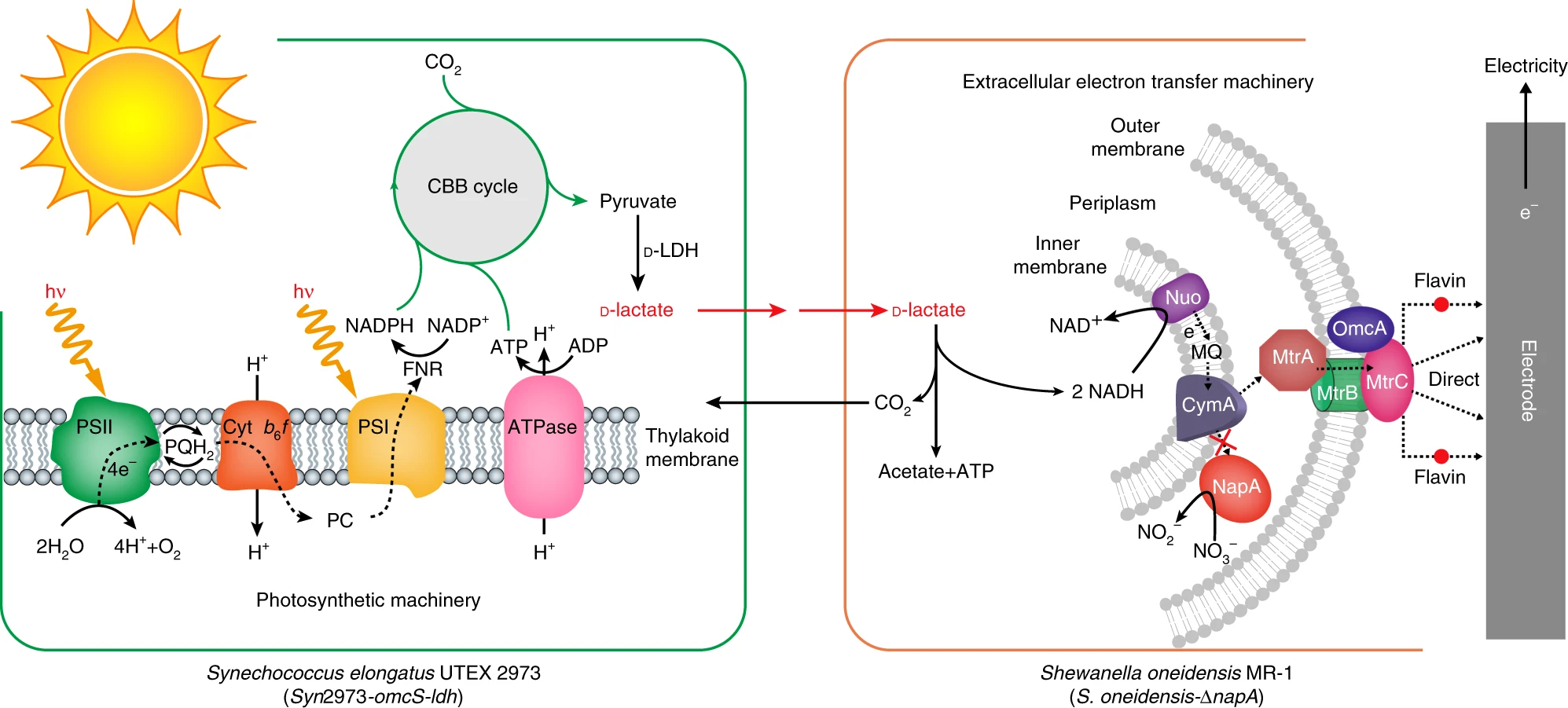
Now comes the (next) challenge - how can these two species be cultured together? the researchers first started with mono-cultures of each strain, making sure each performs their role seperately and efficiently. Now begins a process of manual trial and error. First, they just tried to grow both strains in the cyanobacterial medium, but that didn’t work (no current!). Then they identified a few salts from the second medium that are likely to be critical, and added those. Alas, it didn’t work (no current..). Then they identified two components from the first medium which are probably harmful for the second strain. They removed one, but couldn’t remove the latter since it was critical for the first strain. Then, they engineered the second strain to have a deletion mutation which caused the second strain to longer be affected by this substrate. With this modified medium and engineered strains, the culture grows!
That’s not the end… despite the medium being suitable for both species, the system still did not produce current under light. They figured the reson - the second strain (in charge of discharging) was sensitive to light. So the second innovation that allows this system to work - they created spatial-temporal organization. As can be seen in the following figure:
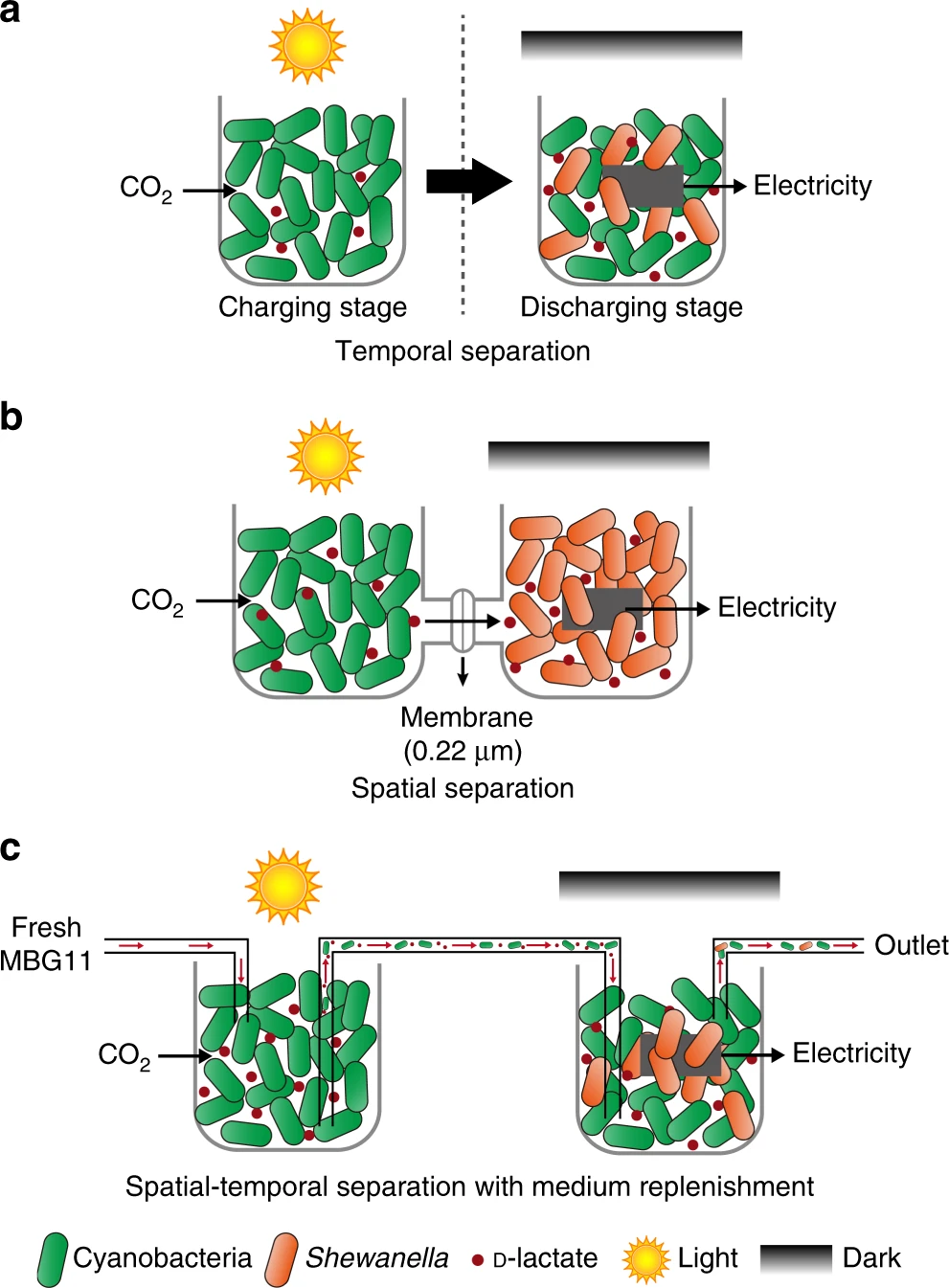
Meaning, the photosynthetic cyanobacterium is exposed to light, while the light-sensitive Shewanella is hidden. The two chambers are connected with the common medium flowing between. The first strain can still pass on the D-lactate, and thus generate electricity. Their system produces electricty in voltage comparable to the best to date (30% than the top recorded BPV). But they main contribution lies in durability - they ran this system for 40 days with a constant electricty generation, while most BPV systems to date decay quickly and a re rarely tested beyond a few days. Pretty cool.
To conclude, in order two cultivate two microbial strains, the researchers had to manually add and subtract ingredients from both mediums, as well as genetically engineer one of the strains. Moreover, a bi-chamber is designed to allow each strain to have it optimal needs for the task in hand.
In my opinion, this method, being manual and knowledge based, would not work well for more than 2 strains or for strains which are not well understood.
Propose a technology for culturing 2 or more strains. How might you innovate in this area given the paper you reviewed?
I am curious to explore technologies that automate the process of medium selection. In the aforementioned paper, the researchers used their logic, knowledge and manual tests to configure each medium, by adding or removing “ingerdients” until they found the right one. What if we build a system which performs a massively parallel screening of co-culture mediums?
The basic idea is that in each microwell, there would be a different mixture of mediums, along with medium mutations. These changes to the medium could be somehow identified, so we could know for each microwell what is exactly there. Then our co-cultures is flowed on the device, and we test what grows and what dies.
This idea is very similar to the method proposed in Massively parallel screening of synthetic microbial communities (Kehe et al, PNAS, 2019), as can be seen in the following figure:
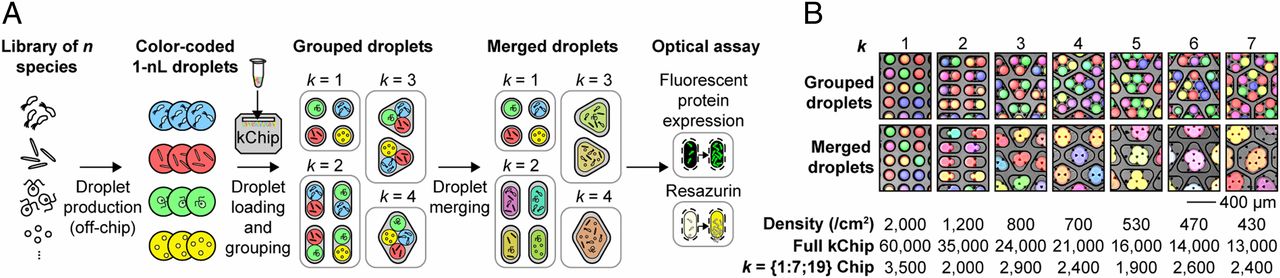
However, in this method, the species are being mixed and match, while I propose we could do the same with a constant number of species, but parallize the process of medium mutations.
It should be noted, that this system does not solve the problem which the above researchers managed to solve manually. While they did “mutate” the mediums, they also mutated the strains themselves to fit the medium. Perhaps, once the above system is validated, an extra step could be an even more multiplexed system, where both microbes and mediums are mutated. Food for thought.
One of the great challenges in microbiology currently is culturing “unculturable” microbes. Propose a methodology for how you might explore this significant space of uncultured microbes.
TBD
Review an article on an artificial gut-on-a-chip technology. What scientific hypotheses are they testing with this in vitro tool? Could you propose an upgrade or innovation to their technique to enable the exploration of other scientific hypotheses? Provide an example of at least one hypothesis you would explore with your proposed system.
In this paper, the researchers aimed to expand the work done on Organs-on-Chips, focusing on the small intestine, and more importantly - using biopsy derived cells. Previous chips used cell lines such as Caco-2, which are very useful for research but contain a lot of mutations and differences compared to cells in vivo. The researchers present a methodology to co-cultivate cells in a microfluidic chamber, and compare it to existing technologies.
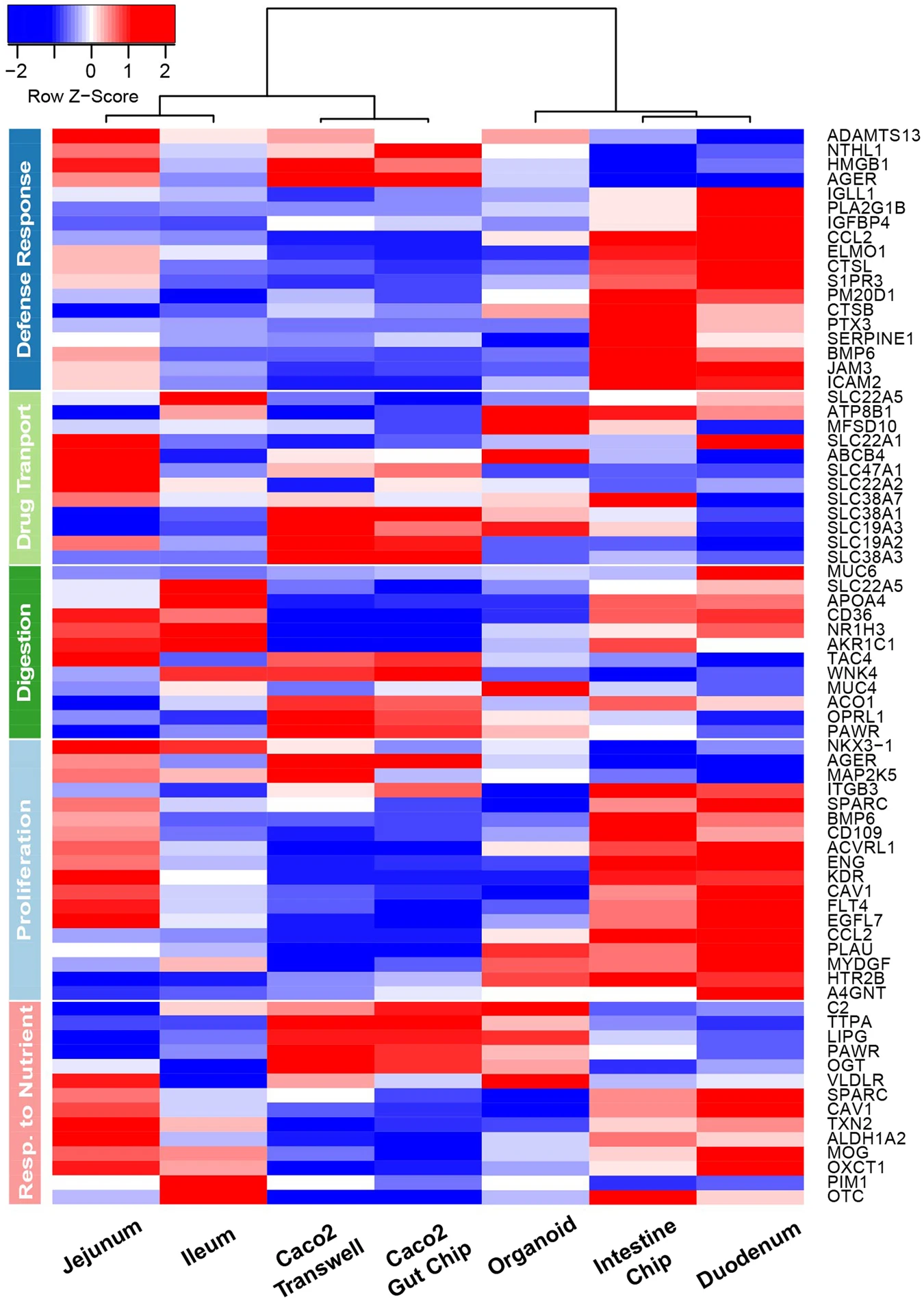
Interestingly, in the above figure you can see on the rightmost column a transcription profile of human native duodenum - which is most similar to the proposed intestine-on-chip (second right column).
In my opinion, these kind of technology is super exciting for precision medicine, an approach where a treatment is being tailored to a specific patient. If we have organ-on-chip systems that can be customized from a specific patient - we can test treatments, drugs etc and look for adverse affects (or no effects) without even giving it to the patient. Precision medicine is already used in some few cases, but the potential is vast and impactful.
In terms of upgrade, as we learned in class, a person’s gut isn’t characterized just by the genetic and transcriptomic profile of its cells, but also (and even, more so) by the microbiota within. Therefore, to create a complete precision medicine solution, we would have to create a intestine-on-chip system which contains both patient-derived cells AND patient-derived microbiota. How to co-culture such a thing is surely a challenge, but a worthy one to pursue.
One of the biggest challenges in public health is quickly detecting new disease outbreaks. How would you go about adaptively responding to new outbreaks? Biobots is using qPCR, so you need to know what you are looking for. How might you develop a technology with a more general view? Some example approaches include microfluidics and point-of-care sequencing, but what else? In particular, are there ways to look for RNA viruses like the flu and SARS-CoV2?
As mentioned, qPCR as well as CRISPR and any approach that is based on knowing a specific sequence will limit us to viruses that we already know about. We could try to find motifs and sequences that are common amongst many viruses. But could we develop a high throughput technology to detect viruses with (almost) zero prior knowledge? if we take some inspiration from the methods mentioned above, organ-on-chips - gut-on-chip, lung-on-chip, etc… what if we had a continuously running organ-on-chip, with an indicator for a viral attack.
Imagine a lung-on-a-chip where wastewater are flowing continuously through. The chip is designed to be resistant to all of the “normal” metabolites and microbiota flowing through. However when a new virus breaks, e.g. SARV-CoV-2, the virus will infect the epithelial cells on the chip and multiply, just like in a human system. We could design some indicator that measures the cell disruption and signal once it passes a certain threshold.